Genetics papers can be pretty impenetrable to us non-geneticists. But geneticists don't write the way they do just so they'll be perceived as eggheads.
It took me, an English major with not much science background beyond high school courses, many months to learn how to decipher the genetics papers that inform Spider Silk. Of course, I was lucky enough to have Cay Craig guide me through these papers and steer me back on track when I veered astray. During this process, I came to realize that genes and genetic research is even more complicated than most of us non-biologists realize. For many of us, it's a mystery why news reports about exciting discoveries in genetics don't lead rapidly to successful medical or other practical applications.
I now get that it's no mystery, or conspiracy. This longer-than-usual post is an attempt to walk through an intriguing paper by a genetics team that writes unusually clearly. Even so, the paper is shot through with terms such as "paralog," "diploid," "retroposition," and "fluorescence in situ hybridization." These terms immediately convey images and lines of logic to other geneticists but gaping black holes to the rest of us. I'm going to avoid such terms as much as possible as I walk through the paper. But I think you'll still see how many interlocking and complex concepts and techniques evolutionary geneticists have to wrestle with, and why even dazzling genetics papers usually lead to more papers rather than to immediate, dramatic applications.
Two of the most closely related spider silk proteins are MaSp1 and MaSp2, proteins that work together in the superstrong, supertough dragline silk of orb-weaving spiders and their closest relatives. Dragline silk is the silk most spiders use as rappelling rope and many also use it as the framing in their webs. MaSp2 protein isn't as strong as MaSp1 protein, but it's stretchier. Having done extensive analyisis, researchers are confident that the MaSp2 gene evolved from the MaSp1 gene. The first spiders to add stretchiness to already-strong dragline silk would have gained an advantage: their web struts could act like shock absorbers, helping their makers cope better with prey impact. So the question isn't why natural selection might have favored such a gene change. It's how the change might have occurred and when. Yonghui Zhao, Nadia Ayoub, and Cheryl Hayashi published a paper last month that announces a new step forward in this quest but also explains how much more spiders still have to teach us.
The quest for the origins of the dragline silk proteins is already decades old. Researchers first discovered sequences for both MaSp1 and MaSp2 in the early 1990s, in Nephila clavipes, a golden orb weaver. These findings led researchers to home in on the sequences of the two proteins and their genes in other spider species, including species in other spider families, the classification above genus.
But these findings were all sequence fragments. Spider silk protein and gene sequences are long and extremely repetitive, which makes coming up with the entire string of amino acids (the units making up proteins) or nucleotides (the units making up genes) challenging. For many years, researchers had to be satisfied with these fragments. They contained enough information to indicate how certain sub-sequences gave rise to dragline silk's superstrength and also to indicate that the dragline proteins in different species all descended from a common ancestor, but fragments are always, well, fragmentary and unsatisfying. What's missing? And does it matter? It took nearly 20 years, until 2007, before a team of researchers (which included Ayoub and Hayashi) pinned down the sequences of full-length MaSp1 and MaSp2 genes, this time in Latrodectus hesperus, the Western black widow. Around the same time, several different research teams found that several different spider species possessed multiple copies of the MaSp1 gene. Natural selection may have favored spiders possessing more than one copy of the MaSp1 gene because they could help spiders produce more MaSp1 protein.
More than 11,000 spider species make both MaSp1 and MaSp2 proteins, so researchers need to think of more efficient research strategies than studying the proteins in every species. Zhao, Ayoub, and Hayashi wanted to see whether the specific location of spider genes within a whole spider genome (which contains thousands of different genes) could tell us anything about how and when MaSp2 evolved from MaSp1. And if the genes' location was interesting, would comparison with the same genes' location on the genome of a different spider species tell us anything more?
One way to find the location of MaSp1 and MaSp2 would be to match their sequences with sub-sequences found within the entire genome sequence. But no one has yet sequenced an entire spider genome. So the team instead set out to locate the genes on chromosomes, which are the packages of DNA that make up the entire genome.
They first determined the karyotype--or number and appearance of the chromosomes--for two spider species, Latrodectus hesperus (Western black widow) and Latrodectus geometricus (brown widow). (What may be interesting about these karyotypes for non-arachnologists is that, unlike humans and most other mammals, spiders don't adhere to the XX-equals-female, XY-equals-male formula. Different spider species have different numbers of chromosomes (20 to 30) and in some species XXXX equals female while XX equals male; in other species, six Xs equal female; one or three Xs equal male in other species. There is a Y chromosome in at least one species of jumping spider. If you'd like to know more about spider reproduction, take a look at Rainer Foelix's definitive Biology of Spiders, which will appear in a new edition in November.)
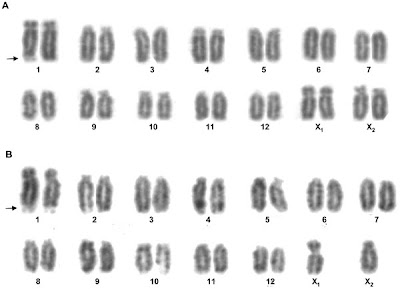
Karyotypes of Latrodectus hesperus, the Western black widow. A is the female karyotype and B is the male karyotype.
Zhao, Ayoub, & Hayashi 2010
Using fluorescent probes that bind only to stretches of DNA that share a highly similar sequence with each probe, the team found that although the multiple MaSp1 genes all clustered closely on one arm of one chromosome, the MaSp2 gene was located on a different chromosome. This finding was the same for both widow species. There was a difference between the species, though: which chromosome the genes were located on and where along the chromosomes they were located. According to the researchers, this difference indicates that the sequences of the two species' genomes have rearranged over the time since the species diverged from their most recent common ancestor, yet the clusters have remained clustered.
There are 31 species in the Latrodectus genus. The Western black widow and the brown widow are very distant cousins within the genus. Yet both have a tight cluster of three MaSp1 genes and a single MaSp2 gene on a different chromosome. This similarity leads to a few conclusions. First, it's probable that the MaSp1 gene multiplied before the Western black widow and the brown widow went their separate ways; that is, their common ancestor already had multiple copies of this gene. If it turns out that the multiple MaSp1 genes already found in two other genera are similarly clustered, the multiplication event or events would be pushed back even farther in time, perhaps as long as about 200 million years ago, when it's estimated these three genera diverged. Having multiple MaSp1 copies must be important to these spiders, because natural selection appears to have favored their continued presence in spiders that aren't very closely related. And if it turns out that they're clustered in all three genera, there's something about that clustering--not just the number-- that's important to individual spider's survival.
Second, these results would be consistent with the hypothesis that the multiplication of the MaSp1 gene resulted from a process known as tandem duplication, which can easily occur as sperm and egg cells are created. (We give a primer on how tandem duplication can occur--and why spider silk genes are so susceptible to this kind of gene change--in Spider Silk.)
And third, they would be consistent with the hypothesis that the MaSp2 gene evolved from the MaSp1 gene. If an MaSp1 copy came unmoored from its fellow copies during some ancient spider's reproductive process and landed at a different location along the genome, it would be exposed to different local genomic sub-sequences from its fellow copies every time sperm or egg cells were created. This would explain both the strong similarities and the marked differences between MaSp1 and MaSp2.
Zhao, Ayoub, and Hayashi appear to be the first researchers to carry out comparative mapping of spider genomes. Similar comparisons carried out on the chromosomes of spiders up and down and across the spider family tree might eventually tell us a lot about the timeline of the evolution of the spider silk gene family and about the selective pressures that drove this evolution. It might also tell us something about the whys and hows of gene clusters, which are found in many different genomes, including humans'. As Cay and I keep repeating, because the trail from spider silk genes to spider silk protein structures to the function of silk proteins is relatively short, the lessons learned from mapping this trail can help us understand even more complex problems in evolution. But it will be quite a few years before we have answers to all our questions about spiders.
It took me, an English major with not much science background beyond high school courses, many months to learn how to decipher the genetics papers that inform Spider Silk. Of course, I was lucky enough to have Cay Craig guide me through these papers and steer me back on track when I veered astray. During this process, I came to realize that genes and genetic research is even more complicated than most of us non-biologists realize. For many of us, it's a mystery why news reports about exciting discoveries in genetics don't lead rapidly to successful medical or other practical applications.
I now get that it's no mystery, or conspiracy. This longer-than-usual post is an attempt to walk through an intriguing paper by a genetics team that writes unusually clearly. Even so, the paper is shot through with terms such as "paralog," "diploid," "retroposition," and "fluorescence in situ hybridization." These terms immediately convey images and lines of logic to other geneticists but gaping black holes to the rest of us. I'm going to avoid such terms as much as possible as I walk through the paper. But I think you'll still see how many interlocking and complex concepts and techniques evolutionary geneticists have to wrestle with, and why even dazzling genetics papers usually lead to more papers rather than to immediate, dramatic applications.
Two of the most closely related spider silk proteins are MaSp1 and MaSp2, proteins that work together in the superstrong, supertough dragline silk of orb-weaving spiders and their closest relatives. Dragline silk is the silk most spiders use as rappelling rope and many also use it as the framing in their webs. MaSp2 protein isn't as strong as MaSp1 protein, but it's stretchier. Having done extensive analyisis, researchers are confident that the MaSp2 gene evolved from the MaSp1 gene. The first spiders to add stretchiness to already-strong dragline silk would have gained an advantage: their web struts could act like shock absorbers, helping their makers cope better with prey impact. So the question isn't why natural selection might have favored such a gene change. It's how the change might have occurred and when. Yonghui Zhao, Nadia Ayoub, and Cheryl Hayashi published a paper last month that announces a new step forward in this quest but also explains how much more spiders still have to teach us.
The quest for the origins of the dragline silk proteins is already decades old. Researchers first discovered sequences for both MaSp1 and MaSp2 in the early 1990s, in Nephila clavipes, a golden orb weaver. These findings led researchers to home in on the sequences of the two proteins and their genes in other spider species, including species in other spider families, the classification above genus.
But these findings were all sequence fragments. Spider silk protein and gene sequences are long and extremely repetitive, which makes coming up with the entire string of amino acids (the units making up proteins) or nucleotides (the units making up genes) challenging. For many years, researchers had to be satisfied with these fragments. They contained enough information to indicate how certain sub-sequences gave rise to dragline silk's superstrength and also to indicate that the dragline proteins in different species all descended from a common ancestor, but fragments are always, well, fragmentary and unsatisfying. What's missing? And does it matter? It took nearly 20 years, until 2007, before a team of researchers (which included Ayoub and Hayashi) pinned down the sequences of full-length MaSp1 and MaSp2 genes, this time in Latrodectus hesperus, the Western black widow. Around the same time, several different research teams found that several different spider species possessed multiple copies of the MaSp1 gene. Natural selection may have favored spiders possessing more than one copy of the MaSp1 gene because they could help spiders produce more MaSp1 protein.
More than 11,000 spider species make both MaSp1 and MaSp2 proteins, so researchers need to think of more efficient research strategies than studying the proteins in every species. Zhao, Ayoub, and Hayashi wanted to see whether the specific location of spider genes within a whole spider genome (which contains thousands of different genes) could tell us anything about how and when MaSp2 evolved from MaSp1. And if the genes' location was interesting, would comparison with the same genes' location on the genome of a different spider species tell us anything more?
One way to find the location of MaSp1 and MaSp2 would be to match their sequences with sub-sequences found within the entire genome sequence. But no one has yet sequenced an entire spider genome. So the team instead set out to locate the genes on chromosomes, which are the packages of DNA that make up the entire genome.
They first determined the karyotype--or number and appearance of the chromosomes--for two spider species, Latrodectus hesperus (Western black widow) and Latrodectus geometricus (brown widow). (What may be interesting about these karyotypes for non-arachnologists is that, unlike humans and most other mammals, spiders don't adhere to the XX-equals-female, XY-equals-male formula. Different spider species have different numbers of chromosomes (20 to 30) and in some species XXXX equals female while XX equals male; in other species, six Xs equal female; one or three Xs equal male in other species. There is a Y chromosome in at least one species of jumping spider. If you'd like to know more about spider reproduction, take a look at Rainer Foelix's definitive Biology of Spiders, which will appear in a new edition in November.)

Karyotypes of Latrodectus hesperus, the Western black widow. A is the female karyotype and B is the male karyotype.
Zhao, Ayoub, & Hayashi 2010
Using fluorescent probes that bind only to stretches of DNA that share a highly similar sequence with each probe, the team found that although the multiple MaSp1 genes all clustered closely on one arm of one chromosome, the MaSp2 gene was located on a different chromosome. This finding was the same for both widow species. There was a difference between the species, though: which chromosome the genes were located on and where along the chromosomes they were located. According to the researchers, this difference indicates that the sequences of the two species' genomes have rearranged over the time since the species diverged from their most recent common ancestor, yet the clusters have remained clustered.
There are 31 species in the Latrodectus genus. The Western black widow and the brown widow are very distant cousins within the genus. Yet both have a tight cluster of three MaSp1 genes and a single MaSp2 gene on a different chromosome. This similarity leads to a few conclusions. First, it's probable that the MaSp1 gene multiplied before the Western black widow and the brown widow went their separate ways; that is, their common ancestor already had multiple copies of this gene. If it turns out that the multiple MaSp1 genes already found in two other genera are similarly clustered, the multiplication event or events would be pushed back even farther in time, perhaps as long as about 200 million years ago, when it's estimated these three genera diverged. Having multiple MaSp1 copies must be important to these spiders, because natural selection appears to have favored their continued presence in spiders that aren't very closely related. And if it turns out that they're clustered in all three genera, there's something about that clustering--not just the number-- that's important to individual spider's survival.
Second, these results would be consistent with the hypothesis that the multiplication of the MaSp1 gene resulted from a process known as tandem duplication, which can easily occur as sperm and egg cells are created. (We give a primer on how tandem duplication can occur--and why spider silk genes are so susceptible to this kind of gene change--in Spider Silk.)
And third, they would be consistent with the hypothesis that the MaSp2 gene evolved from the MaSp1 gene. If an MaSp1 copy came unmoored from its fellow copies during some ancient spider's reproductive process and landed at a different location along the genome, it would be exposed to different local genomic sub-sequences from its fellow copies every time sperm or egg cells were created. This would explain both the strong similarities and the marked differences between MaSp1 and MaSp2.
Zhao, Ayoub, and Hayashi appear to be the first researchers to carry out comparative mapping of spider genomes. Similar comparisons carried out on the chromosomes of spiders up and down and across the spider family tree might eventually tell us a lot about the timeline of the evolution of the spider silk gene family and about the selective pressures that drove this evolution. It might also tell us something about the whys and hows of gene clusters, which are found in many different genomes, including humans'. As Cay and I keep repeating, because the trail from spider silk genes to spider silk protein structures to the function of silk proteins is relatively short, the lessons learned from mapping this trail can help us understand even more complex problems in evolution. But it will be quite a few years before we have answers to all our questions about spiders.